Introduction
This project aims at performing automated identification of cell boundaries from the pathological video data.
We are given the video file cells.avi as input. The problem statement can be found here.
The input video file:
Compatibility
- This code has been tested on Ubuntu 16.04 LTS and Windows 10
- Dependencies - Python 2.7 & 3.5, OpenCV 3.0+
Methods Used
- Image Processing followed by Contours
- Adaptive Thresholding
- Watershed Algorithm
- Structured Forest Algorithm
Usage
First clone the repository by typing: git clone https://github.com/iitmcvg/Cell-Segmentation.git.
Structured Forest
- First execute
python framesaver.pyto save the frames for structured forest. - Next execute
python StructuredForests.pyto apply the edge detection. - Finally, execute
python videowriter.pyto write the outputs to a video file.
Results
- The video
edge.aviis the result after applying Structured Forest algorithm. Other outputs can be found in theOutputsfolder. - Outputs of all methods can be seen at once in this video.
The output video file:
We include a few output screenshots here:

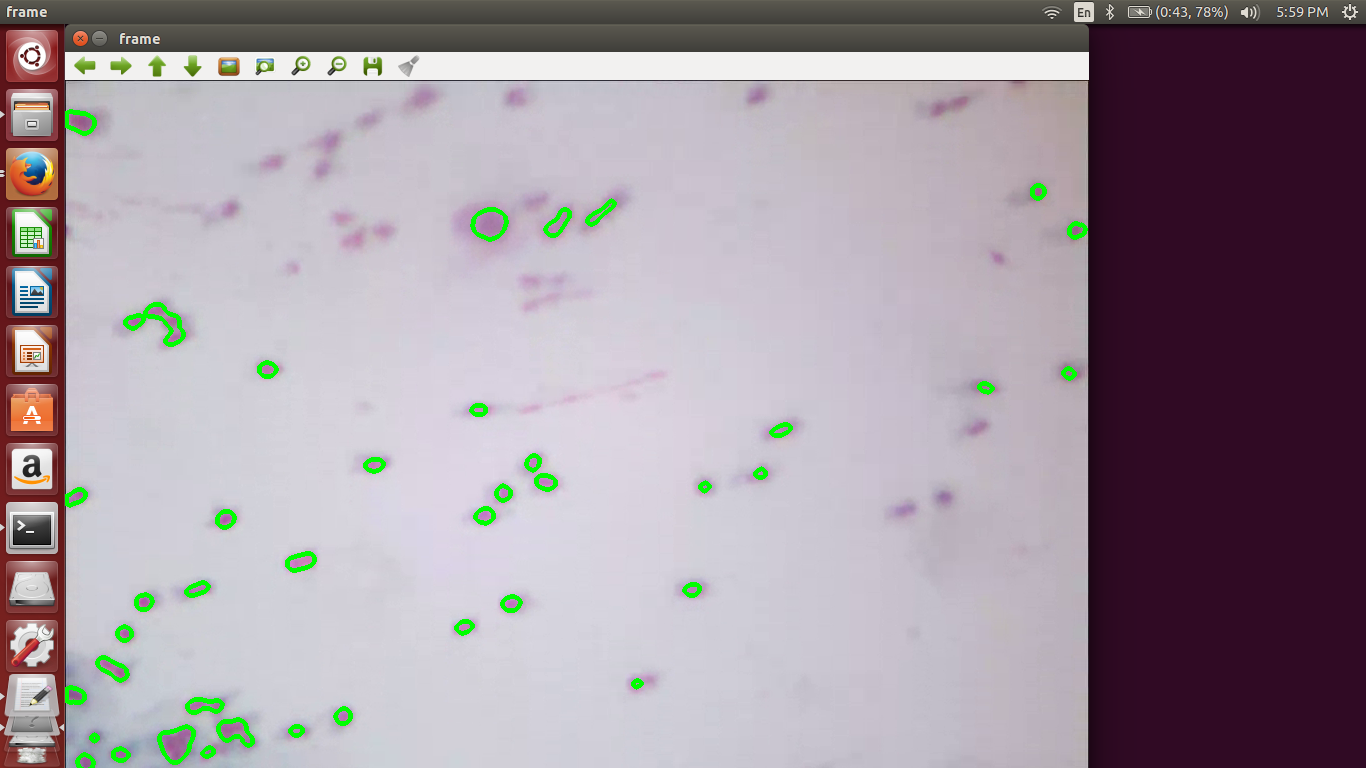
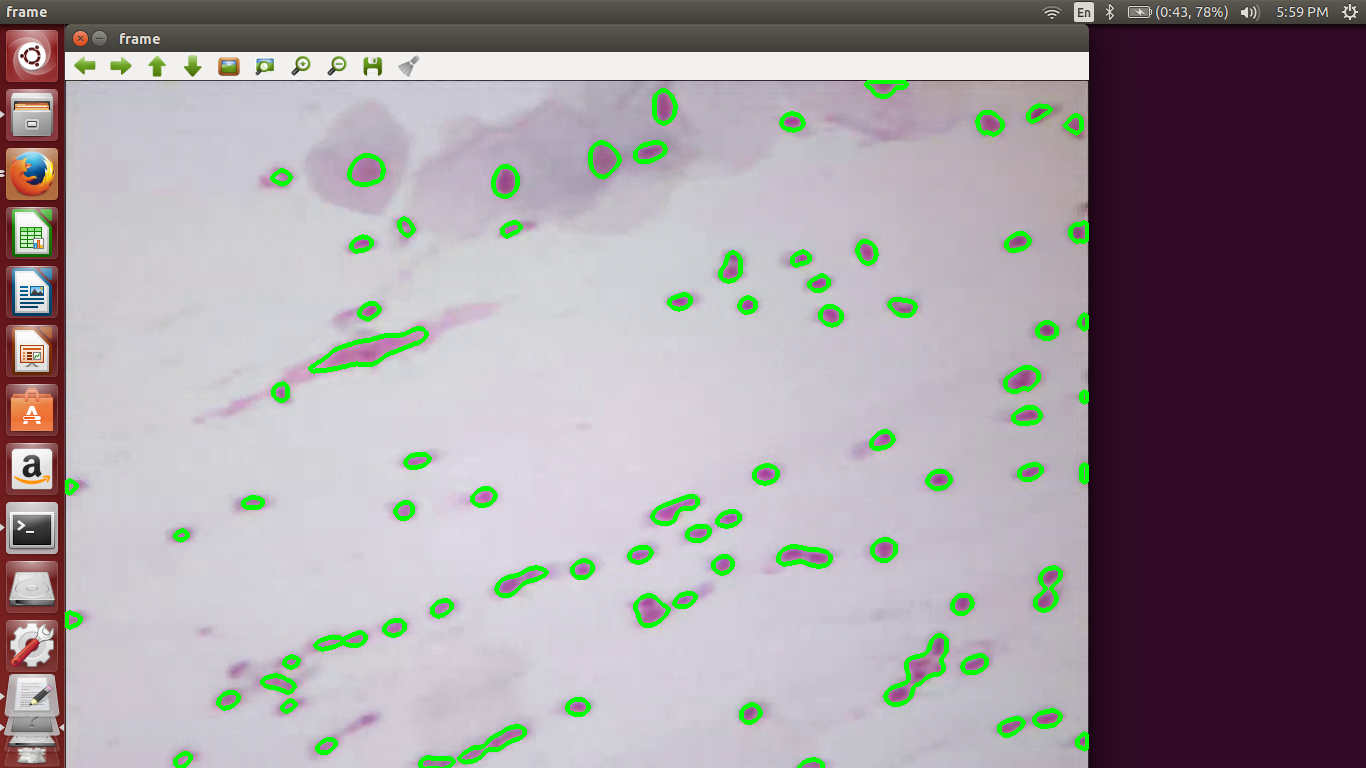
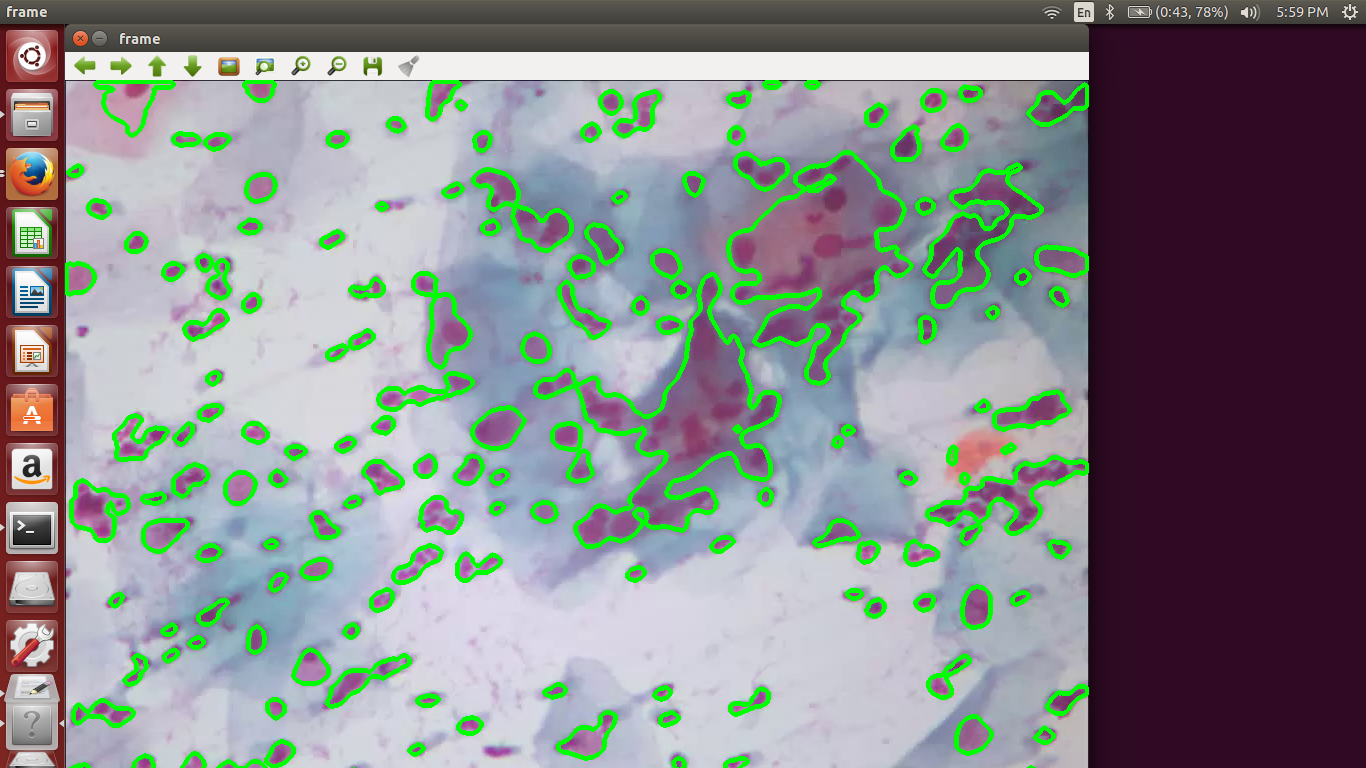
References
Our Structured Forest is an implementation of Artanis CV Structured Forest.
Future work
-
U-net convolutional neural network can be used.
-
Implementing the algorithm given in this paper.
Disclaimer
This software is published for academic and non-commerical use only.




Leave a Comment